Accessing Sequence Data via NCI’s Cancer Data Service (CDS)
!!! NOTE: dbGaP approval for HTAN study phs002371 is required in order to access HTAN lower-level genomics data, such as RNAseq FASTQ and BAM files. !!!
The CDS Portal, within NCI’s Cancer Research Data Commons (CRDC), provides an interface to filter and select data from a variety of NCI programs, including controlled-access, primary sequence data from the Human Tumor Atlas Network (HTAN). This page provides directions for importing sequencing data from CDS to the Cancer Genomics Commons (CGC).
The directions for accessing sequencing data on CDS are similar to those for Level 2 Imaging Data Access, including Direct Export from CDS to CGC and importing data using a Data Repository Service (DRS) Manifest. Please follow the Level 2 Imaging Data Access directions to access sequencing data, noting the following changes:
- For Direct Export or Generating a DRS Manifest from CDS, choose Human Tumor Atlas (HTAN) primary sequence data on the STUDY section of the left hand sidebar instead of Human Tumor Atlas (HTAN) imaging data.
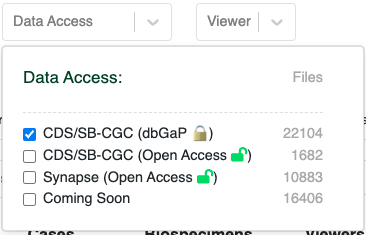
- To generate a DRS Manifest from the HTAN Data Portal, click CDS/SB-CGC (dbGaP) under the Data Access filter instead of CDS/SB-CGC (Open Access).