#
Relationship Model
HTAN Centers have multiple research participants, who have donated biospecimens (see Figure below). Metadata submitted by HTAN Centers allows one to trace back any data file to the donated biospecimen. Data may be derived directly from the original biospecimen or from subsamples/assayed samples. Via metadata, Level 1 raw data files are often directly linked to the corresponding assayed biospecimen, whereas processed Level 2-4 data files are linked to lower level parent data files. Connecting a given data file to the originating biospecimen often requires following the ID relationships (ID Provenance) contained within metadata.

#
HTAN ID Provenance Table
The HTAN DCC has constructed an ID Provenance BigQuery table, which contains upstream biospecimen and participant information for each HTAN data file. The motivation for constructing the table arose from the need for a straightforward method of showcasing how HTAN data files are linked to biospecimens and patients.
Although this information is available in HTAN, connections among assay file levels and parent relationships (both biospecimen and file) were previously only accessible by tracing through parent identifiers. Having all ID information in one place can significantly speed up analyses, exploration, and data sharing.
The Provenance BigQuery table is accessible via ISB-CGC. See the Google BigQuery section for more details.
#
Biospecimen Attribute Definitions
Given the complexity of biospecimen relationships, we've adopted the following nomenclature to describe biospecimen lineages:
Originating Biospecimen: the biopsied or resected biospecimen from the patient from which the assay data were derived
Assayed Biospecimen: the biospecimen directly assayed using the experimental platform
Biospecimen Path: path of biospecimens from Originating to Assayed; comma-separated
#
Provenance Table Construction
As illustrated in the figure above, biospecimens can be subsampled multiple times. However, HTAN metadata tables provide only the immediate parent biospecimen. To assemble the full biospecimen path, we perform a series of joins on the biospecimen table, walking up the parent biospecimen ancestry chain until no further parents are found.
Similarly, we can have up to four data file ‘levels’. Each data file is linked using its provided parent HTAN data file ID(s).
We then join biospecimen information with file-level annotations to form the final ID provenance table.
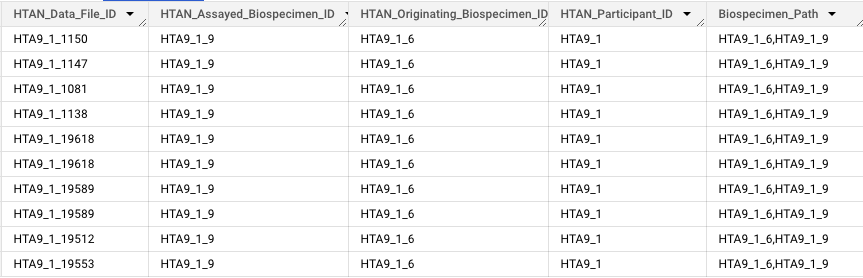
Please note: In HTAN Phase 1, Biospecimen and Data File IDs are numeric only. In Phase 2, Biospecimen IDs include a "B" and Data File IDs include a "D". Please see the identifiers page for more information.
#
Table Contents
In addition to upstream biospecimen and participant IDs, the provenance table also includes a number of informational columns, such as entityId (Synapse ID of the source file), HTAN_Center (text version of center code), and Data_Release and CDS_Release which indicate which HTAN Portal release and CDS release the file was included in, respectively.
